FATE tutorial 2 - Run a simulation
Source:vignettes/fate_tutorial_2_RUN_SIMULATION.Rmd
fate_tutorial_2_RUN_SIMULATION.Rmd“The recurring suggestions are that models should explicitly (i) include spatiotemporal dynamics; (ii) consider multiple species in interactions and (iii) account for the processes shaping biodiversity distribution.”
FATE is a “a biodiversity model that meets this
challenge at regional scale by combining phenomenological and
process-based approaches and using well-defined plant functional
group”. (Boulangeat,
2014)
What FATE needs to be run?
The different type of parameters and flags
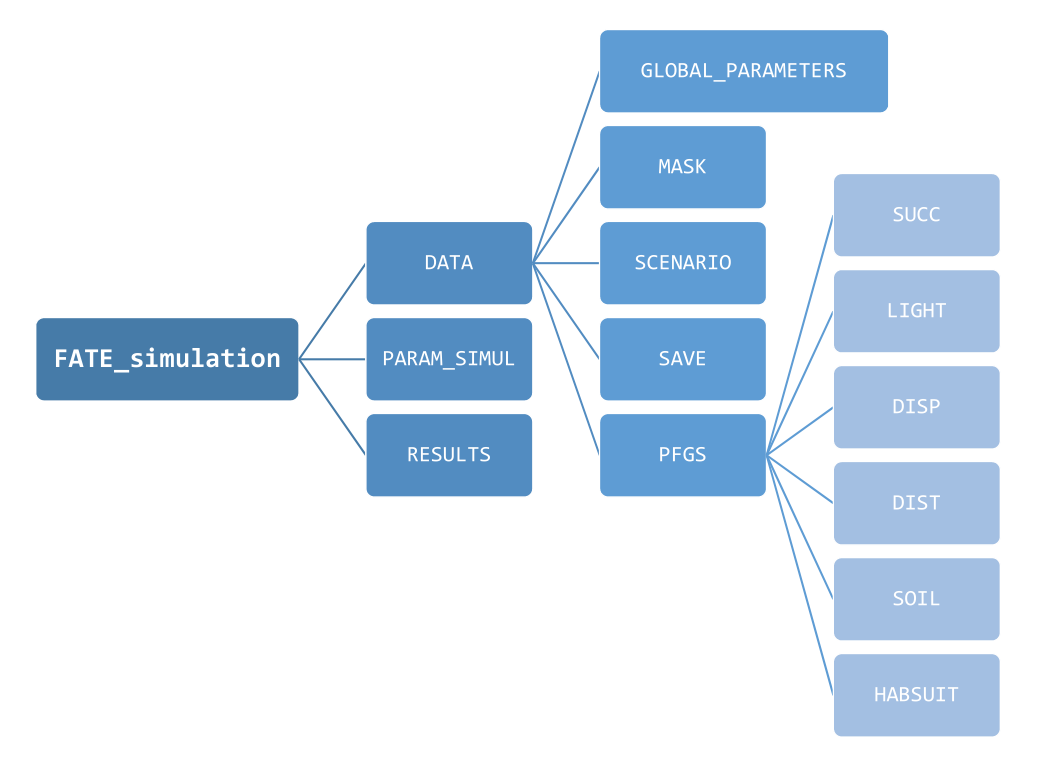
FATE requires a quite large number of parameters, which
are stored into .txt files, presented to and recovered by
the software. These parameters can be of 3 types :
- Filenames, to guide the application to other parameter files that should be read
- These filenames either correspond to :
- other parameter files that contain values to be actually read and used
-
raster files, with the extension
.tif(lighter) or.img
How to produce all these parameters files ?
RFate is a R package available on github and designed to
provide support function to the FATE software.
It contains documentation and functions to create and organize all input files required by the model, as well as scripts to help analyze the resulting outputs.
RFate tutorial
The FATE friendly directory organization
As FATE needs quite a lot of parameters given into
different parameter files, it is important to organize them in a clear
and intuitive way.
While this is not mandatory to run a FATE simulation, a
specific and methodical folder architecture can be obtained with the PRE_FATE.skeletonDirectory
function from the RFate package.
A good way to start is to create this skeleton directory, and then copy or create the needed files inside to have all simulation data into the same place.
How to run a FATE Simulation ?
When all data and parameter files have been produced and correctly
referred in a SimulParameters
file, a FATE simulation can be run using the FATE function :
- the simulation will start, and the software will print messages into the console indicating what the software is doing.
- depending on the simulation duration, the size and the resolution of the study area, the number of Plant Functional Groups (PFG) involved, … running the full simulation could take a while.
Note : the folder from which the command is sent must be adapted based on how paths to files have been given within the SimulParameters file :
If all paths are absolute
(i.e. including the root, such as/home/username/FATE_simulation/DATA/GLOBAL_PARAMETERS/Global_parameters_V1.txt),
there should not be any problem.
The only requirement then is to also give absolute path to the simulation folder, if not in the current directory.If all paths are relative
(i.e. starting from a specific folder, such asFATE_simulation/DATA/GLOBAL_PARAMETERS/Global_parameters_V1.txt),
then theFATEsimulation must be run from this specific folder
(i.e. here from the folder containing theFATE_simulationfolder).
How to handle the results ?
Once the simulation is completed, the directory defined under the
flag --SAVING_DIR-- within the SimulParameters
file can contain the following directories (depending on the parameters
selected and the modules activated within the GlobalParameters
file) :
ABUND_allPFG_perStrata/ABUND_perPFG_allStrata/ABUND_perPFG_perStrata/
Each of them contains raster files with the abundance of Plant Functional Groups (PFG) (which should be considered as a proxy for the vegetation coverage/abundance) contained within each pixel of the study area. The files within each folder show different informations :
-
ABUND_perPFG_perStrata/: a file represents one specific year, and the abundance of a specific PFG in a specific stratum -
ABUND_allPFG_perStrata/ : a file represents one specific year, and the abundance of all PFGs in a specific stratum -
ABUND_perPFG_allStrata/: a file represents one specific year, and the abundance of a specific PFG within all strata
If the corresponding modules were activated within the GlobalParameters file, the following folders may also exist and contain output files :
DISPERSAL/LIGHT/SOIL/
These ouputs can then be used as is, or post-treated with functions from the RFate package to obtain more specific results (e.g. evolution of abundance curves, cover or diversity maps, etc).
RFate tutorial